
 Ph.D. Student at Shanghai Jiao Tong University
Ph.D. Student at Shanghai Jiao Tong UniversityI am a first-year Ph.D. student in Electronic Information at the School of Artificial Intelligence (SAI), Shanghai Jiao Tong University (SJTU), advised by Prof. Peilin Zhao. Prior to this, I completed my M.S. degree in Computer Science and Technology at the School of Computing and Artificial Intelligence, Southwest Jiaotong University (SWJTU), under the supervision of Professor Yan Yang and Assistant Researcher Yongquan Jiang. I also received my Bachelor of Science degree in Electronic Information Science and Technology from the School of Science, Southwest Petroleum University (SWPU).
My research lies at the intersection of AI4Science and Molecular Machine Learning, with a particular focus on the development and application of Generative Algorithms.
Warning
Problem: The current name of your GitHub Pages repository ("Solution: Please consider renaming the repository to "
http://".
However, if the current repository name is intended, you can ignore this message by removing "{% include widgets/debug_repo_name.html %}" in index.html.
Action required
Problem: The current root path of this site is "baseurl ("_config.yml.
Solution: Please set the
baseurl in _config.yml to "Education
-
 Shanghai Jiao Tong UniversitySchool of Artificial Intelligence (SAI)
Shanghai Jiao Tong UniversitySchool of Artificial Intelligence (SAI) Ph.D. Student in Electronic InformationSep. 2025 - present
Ph.D. Student in Electronic InformationSep. 2025 - present -
 Southwest Jiaotong UniversitySchool of Computing and Artificial IntelligenceM.S. Student in Computer Science and TechnologySep. 2022 - Jun. 2025
Southwest Jiaotong UniversitySchool of Computing and Artificial IntelligenceM.S. Student in Computer Science and TechnologySep. 2022 - Jun. 2025 -
 Southwest Petroleum UniversitySchool of ScienceB.S. in Electronic Information Science and TechnologySep. 2018 - Jun. 2022
Southwest Petroleum UniversitySchool of ScienceB.S. in Electronic Information Science and TechnologySep. 2018 - Jun. 2022
Experience
-
 Megvii (Chengdu)Research Intern [Cryo-EM Structure Reconstruction]May. 2024 - Jan. 2025
Megvii (Chengdu)Research Intern [Cryo-EM Structure Reconstruction]May. 2024 - Jan. 2025 -
 Tencent AI LabResearch Intern [AI for (Molecule & Materials) Science]Feb. 2025 - Oct. 2025
Tencent AI LabResearch Intern [AI for (Molecule & Materials) Science]Feb. 2025 - Oct. 2025
Honors & Awards
-
2nd Class Academic Scholarship, Southwest Jiaotong University (SWJTU)Oct. 2022
-
2nd Class Academic Scholarship, Southwest Jiaotong University (SWJTU)Oct. 2023
-
1st Class Academic Scholarship, Southwest Jiaotong University (SWJTU)Oct. 2024
-
National Scholarship for M.S., Southwest Jiaotong University (SWJTU)Oct. 2024
News
Selected Publications (view all )
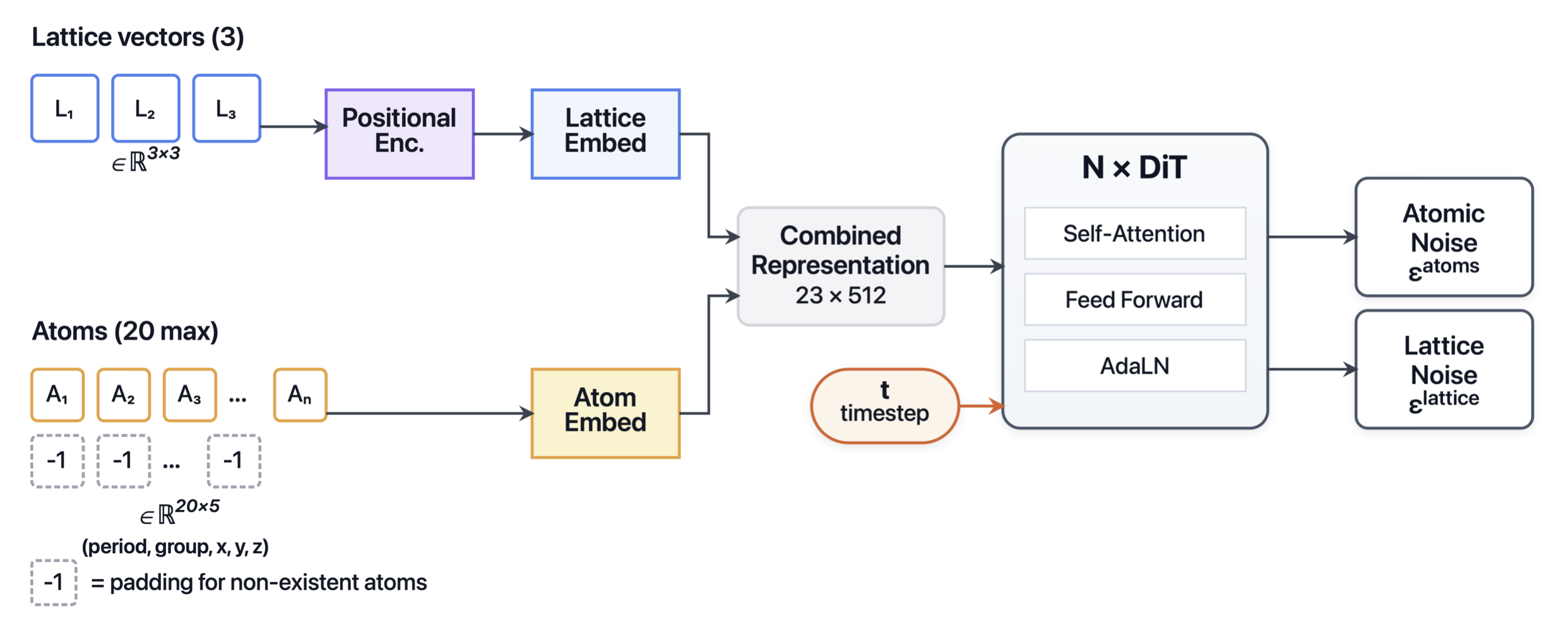
CrystalDiT: Simple Diffusion Transformers for Crystal Generation
Xiaohan Yi, Guikun Xu, Xi Xiao#, Zhong Zhang, Liu Liu, Yatao Bian, Peilin Zhao# (# corresponding author)
The 40th Annual AAAI Conference on Artificial Intelligence AAAI26 2025-10
We present CrystalDiT, a diffusion transformer for crystal structure generation that achieves state-of-the-art performance by challenging the trend of architectural complexity. Instead of intricate, multi-stream designs, CrystalDiT employs a unified transformer that imposes a powerful inductive bias: treating lattice and atomic properties as a single, interdependent system. Combined with a periodic table-based atomic representation and a balanced training strategy, our approach achieves 9.62% SUN (Stable, Unique, Novel) rate on MP-20, substantially outperforming recent methods including FlowMM (4.38%) and MatterGen (3.42%). Notably, CrystalDiT generates 63.28% unique and novel structures while maintaining comparable stability rates, demonstrating that architectural simplicity can be more effective than complexity for materials discovery. Our results suggest that in data-limited scientific domains, carefully designed simple architectures outperform sophisticated alternatives that are prone to overfitting..
CrystalDiT: Simple Diffusion Transformers for Crystal Generation
Xiaohan Yi, Guikun Xu, Xi Xiao#, Zhong Zhang, Liu Liu, Yatao Bian, Peilin Zhao# (# corresponding author)
The 40th Annual AAAI Conference on Artificial Intelligence AAAI26 2025-10
We present CrystalDiT, a diffusion transformer for crystal structure generation that achieves state-of-the-art performance by challenging the trend of architectural complexity. Instead of intricate, multi-stream designs, CrystalDiT employs a unified transformer that imposes a powerful inductive bias: treating lattice and atomic properties as a single, interdependent system. Combined with a periodic table-based atomic representation and a balanced training strategy, our approach achieves 9.62% SUN (Stable, Unique, Novel) rate on MP-20, substantially outperforming recent methods including FlowMM (4.38%) and MatterGen (3.42%). Notably, CrystalDiT generates 63.28% unique and novel structures while maintaining comparable stability rates, demonstrating that architectural simplicity can be more effective than complexity for materials discovery. Our results suggest that in data-limited scientific domains, carefully designed simple architectures outperform sophisticated alternatives that are prone to overfitting..

CoFM: Molecular Conformation Generation via Flow Matching in SE(3)-Invariant Latent Space
Guikun Xu*, Yankai Yu*, Yongquan Jiang#, Yan Yang, Yatao Bian# (* equal contribution, # corresponding author)
Forty-Second International Conference on Machine Learning GenBio Workshop ICML25 GenBio 2025-07
Current leading methods for molecular conformation generation often rely on computationally intensive diffusion models in 3D space, which struggle with accurately modeling conformational manifolds and rigorously maintaining SE(3) equivariance. These limitations hinder both performance and efficiency, and can complicate integration with standard tools like RDKit. To overcome these challenges, we introduce CoFM, a novel generative framework that pioneers the concept of an autoencoder-induced, fully SE(3)-invariant latent space. This approach decouples SE(3) equivariance constraints from the generation process, enabling seamless integration of RDKit’s physicochemical priors. Furthermore, CoFM is the first to integrate latent flow matching within this invariant geometric subspace, significantly enhancing generation efficacy with fewer iterative steps. Experimental validation demonstrates that our method generates high-quality results with fewer iterations, achieving significant improvements in key Precision metrics and ensuring greater energy authenticity.
CoFM: Molecular Conformation Generation via Flow Matching in SE(3)-Invariant Latent Space
Guikun Xu*, Yankai Yu*, Yongquan Jiang#, Yan Yang, Yatao Bian# (* equal contribution, # corresponding author)
Forty-Second International Conference on Machine Learning GenBio Workshop ICML25 GenBio 2025-07
Current leading methods for molecular conformation generation often rely on computationally intensive diffusion models in 3D space, which struggle with accurately modeling conformational manifolds and rigorously maintaining SE(3) equivariance. These limitations hinder both performance and efficiency, and can complicate integration with standard tools like RDKit. To overcome these challenges, we introduce CoFM, a novel generative framework that pioneers the concept of an autoencoder-induced, fully SE(3)-invariant latent space. This approach decouples SE(3) equivariance constraints from the generation process, enabling seamless integration of RDKit’s physicochemical priors. Furthermore, CoFM is the first to integrate latent flow matching within this invariant geometric subspace, significantly enhancing generation efficacy with fewer iterative steps. Experimental validation demonstrates that our method generates high-quality results with fewer iterations, achieving significant improvements in key Precision metrics and ensuring greater energy authenticity.

Cryo-EM Structure Reconstruction by Gaussian Splatting: Pushing the Resolution to Extrem
Shuaicheng Liu, Shen Cheng, Guikun Xu, Haoqiang Fan, Bing Zeng# (# corresponding author)
Submitted to (Under review) Preprint 2025-03
In the field of structural biology, Cryo-EM based high-resolution 3-D structure reconstruction of complex macromolecules is a vital step. Although multiple attempts have been tried within this framework to consider quality-degrading factors such as imaging noise, non-uniform distribution of particle orientations, and sample heterogeneity in order to achieve high resolution, there is still a substantial gap between the best reconstruction resolution achieved by the existing methods and the hard resolution provided by the imaging device. Here, we introduce CryoGS, a novel 3-D reconstruction method for Cryo-EM structures using Gaussian splatting. Through the integration of 3-D Gaussian representations into neural network learning, CryoGS employs a spatial domain approach to optimize learnable 3-D Gaussians and project them into 2-D images using the splatting technique. Compared with the existing methods, CryoGS achieves significant improvements in resolution, isotropy, and computational efficiency. For example, CryoGS achieves a resolution of 2.217 $\AA$ on EMPIAR-10492 dataset, approaching its theoretical limit of 2.2 $\AA$, while the best resolution achieved by the existing methods is 3.805 $\AA$. Furthermore, CryoGS exhibits remarkable robustness in reconstructing heterogeneous structures and high-resolution models under extreme conditions such as pose inaccuracy, limited particle data, and high noise. Based on these results, we believe that CryoGS has great potential to be a powerful tool for Cryo-EM applications to ensure enhanced resolution, robustness, and efficiency.
Cryo-EM Structure Reconstruction by Gaussian Splatting: Pushing the Resolution to Extrem
Shuaicheng Liu, Shen Cheng, Guikun Xu, Haoqiang Fan, Bing Zeng# (# corresponding author)
Submitted to (Under review) Preprint 2025-03
In the field of structural biology, Cryo-EM based high-resolution 3-D structure reconstruction of complex macromolecules is a vital step. Although multiple attempts have been tried within this framework to consider quality-degrading factors such as imaging noise, non-uniform distribution of particle orientations, and sample heterogeneity in order to achieve high resolution, there is still a substantial gap between the best reconstruction resolution achieved by the existing methods and the hard resolution provided by the imaging device. Here, we introduce CryoGS, a novel 3-D reconstruction method for Cryo-EM structures using Gaussian splatting. Through the integration of 3-D Gaussian representations into neural network learning, CryoGS employs a spatial domain approach to optimize learnable 3-D Gaussians and project them into 2-D images using the splatting technique. Compared with the existing methods, CryoGS achieves significant improvements in resolution, isotropy, and computational efficiency. For example, CryoGS achieves a resolution of 2.217 $\AA$ on EMPIAR-10492 dataset, approaching its theoretical limit of 2.2 $\AA$, while the best resolution achieved by the existing methods is 3.805 $\AA$. Furthermore, CryoGS exhibits remarkable robustness in reconstructing heterogeneous structures and high-resolution models under extreme conditions such as pose inaccuracy, limited particle data, and high noise. Based on these results, we believe that CryoGS has great potential to be a powerful tool for Cryo-EM applications to ensure enhanced resolution, robustness, and efficiency.

GTMGC: Using Graph Transformer to Predict Molecule’s Ground-State Conformation
Guikun Xu, Yongquan Jiang#, Pengchuan Lei, Yan Yang, Jim Chen (# corresponding author)
Twelfth International Conference on Learning Representations ICLR24 Spotlight 2024-01
The ground-state conformation of a molecule is often decisive for its properties. However, experimental or computational methods, such as density functional theory (DFT), are time-consuming and labor-intensive for obtaining this conformation. Deep learning (DL) based molecular representation learning (MRL) has made significant advancements in molecular modeling and has achieved remarkable results in various tasks. Consequently, it has emerged as a promising approach for directly predicting the ground-state conformation of molecules. In this regard, we introduce GTMGC, a novel network based on Graph-Transformer (GT) that seamlessly predicts the spatial configuration of molecules in a 3D space from their 2D topological architecture in an end-to-end manner. Moreover, we propose a novel self-attention mechanism called Molecule Structural Residual Self-Attention (MSRSA) for molecular structure modeling. This mechanism not only guarantees high model performance and easy implementation but also lends itself well to other molecular modeling tasks. Our method has been evaluated on the Molecule3D benchmark dataset and the QM9 dataset. Experimental results demonstrate that our approach achieves remarkable performance and outperforms current state-of-the-art methods as well as the widely used open-source software RDkit.
GTMGC: Using Graph Transformer to Predict Molecule’s Ground-State Conformation
Guikun Xu, Yongquan Jiang#, Pengchuan Lei, Yan Yang, Jim Chen (# corresponding author)
Twelfth International Conference on Learning Representations ICLR24 Spotlight 2024-01
The ground-state conformation of a molecule is often decisive for its properties. However, experimental or computational methods, such as density functional theory (DFT), are time-consuming and labor-intensive for obtaining this conformation. Deep learning (DL) based molecular representation learning (MRL) has made significant advancements in molecular modeling and has achieved remarkable results in various tasks. Consequently, it has emerged as a promising approach for directly predicting the ground-state conformation of molecules. In this regard, we introduce GTMGC, a novel network based on Graph-Transformer (GT) that seamlessly predicts the spatial configuration of molecules in a 3D space from their 2D topological architecture in an end-to-end manner. Moreover, we propose a novel self-attention mechanism called Molecule Structural Residual Self-Attention (MSRSA) for molecular structure modeling. This mechanism not only guarantees high model performance and easy implementation but also lends itself well to other molecular modeling tasks. Our method has been evaluated on the Molecule3D benchmark dataset and the QM9 dataset. Experimental results demonstrate that our approach achieves remarkable performance and outperforms current state-of-the-art methods as well as the widely used open-source software RDkit.